Molecular Study of Vancomycin Resistance in Staphylococcus aures associated with Nosocomial Infections - Juniper publishers
Journal of Trends in Technical and Scientific Research
Abstract
Background:Staphylococcus aureus (S. aureus) causes hospital associated infections (HAIs).
Aim:The aim of the present study was to
identify the emergence of vancomycin-resistant S. aureus among MRSA
resistant andto identify the occurrence of van A, van B and van C genes
among resistant isolates.
Method:The isolated strains confirmed to be S.
aureus were subjected to full microbiological laboratory study for
identification and antibiotics susceptibility beside molecular study for
detection of vanA, vanB and van C genes by multiplex PCR.
Results:The study included 365 isolated S.
aureus strains. Among isolated S.aureus strains, 113 (30.9%) was found
to be MRSA. van A gene was recognized among 13 (68. 4%) resistant
strains. van B was more commonly presents among resistant strains
17(89.5%).
Keywords: MRSA Multiplex PCR VanA VanB Vancomycin resistance
Introduction
Staphylococcus aureus is a leading pathogen in
hospital acquired infections. It is isolated from various hospital
acquired infections and its pathogenicity increased with the emergence
of methicillin resistance (MRSA) in the last decades [1]. Vancomycin
antibiotic is a glycopeptide antibiotic which have been considered a
good therapeutic alternative for the treatment of MRSA. Unfortunately,
resistant strains have been reported to reemerge among S. aureus
species. The resistant strains have been reported to acquire thick wall
preventing diffusion of vancomycin to the bacterial cells [2].
Vancomycin-resistant genes associated with S.aureus species are like
those present in Enterococcus spp. These genes are seven types of
resistance genes namely (vanA, B, C, D, E, G, and L). They are usually
transferred from Enterococcus spp, by transposon Tn1546 [3].
The aim of the present study was to identify the
emergence of vancomycin-resistant S. aureus among MRSA resistant strains
and to identify the occurrence of vanA, vanB and van C genes among
resistant isolates.
Materials and Methods
The study is a retrospective observational case
series study that was conducted at Mansoura University Children
hospital, Egypt from December 2014 till March 2016. The study included
isolated S. aureus strains from children diagnosed to have health care
associated infections (HCAI) according to CDC criteria of HCAI [4]. The
patients signed written consents and the study was approved by Mansoura
Faculty of Medicine ethical committee.
The isolated strains confirmed to be S. aureus by
automated identification system Microscan (Bechman, USA), were subjected
to full microbiological laboratory study including antibiotics
susceptibility tests by disc diffusion method, manual determination of
minimal inhibitory concentration for vancomycin and molecular study for
detection of vanA, vanB and vanC genes by multiplex PCR.
Antibiotics susceptibility test
The used discs were vancomycin (30μg), erythromycin
(15μg), ampicillin/sulbactam (20μg), amoxicillin/clavulanic
(20/10μg), clindamycin (5μg)/, ceftriaxone 5μg, ceftazidime
(30μg), cefoperazone (75μg), gentamycin (30μg), cefoxitin disc
(30μg) (Oxoid Hampshire, England). Determination MRSA isolates
was reported as those strains with inhibition zone ≤ 21 mm.
Broth Dilution Method of minimal inhibitory concentrations (MICs) for vancomycin
The determination of minimal inhibitory concentrations
(MICs) for vancomycin was performed using standardized broth
dilution techniques [5].
Vancomycin resistance among MRSA according to MIC was
classified into susceptible, intermediate susceptible and resistant
according to CLSI, 2009 [6].
Multiplex PCR for Van A, B, C genes Determination for MRSA strains
DNA preparation:One colony of pure culture was suspended
in 25μL of sterile water and the suspension was put in the water
bath at 100°C for 12 minutes. One micron of the suspension was
used for PCR amplification.
Multiplex PCR:The primers sequences used in PCR and
amplification were as follow, vanA 5/-ATG AAT AGA ATA AAA GTT
GC-/3, 5/-TCA CCC CTT TAA CGC TAA TA-/3 bp1032 [7], vanB 5/-
GTG ACA AAC CGG AGG CGA GGA-/3, 5/-CCG CCA TCC TCC TGC
AAA AAA-/3, 430bp [8], vanC 5/-ACG AGA AAG ACA ACA GGA AGA
CC-/3, 5/-ACA TCG TGA TCG CTA AAA GGA GC-/3, 815bp [9].
The multiplex PCR was performed according to Perez-Roth
et al. [10] using Qiagen amplification kit. Sterile distilled water
was used as a negative control under complete sterile standard
precautions for PCR.
After amplification 10μL of the reaction mixture was loaded
onto a 1% agarose gel stained with 10μL ethidium bromide and
electrophoresed to estimate the sizes of the amplification products
with a 100-bp molecular size standard ladder (Sigma).
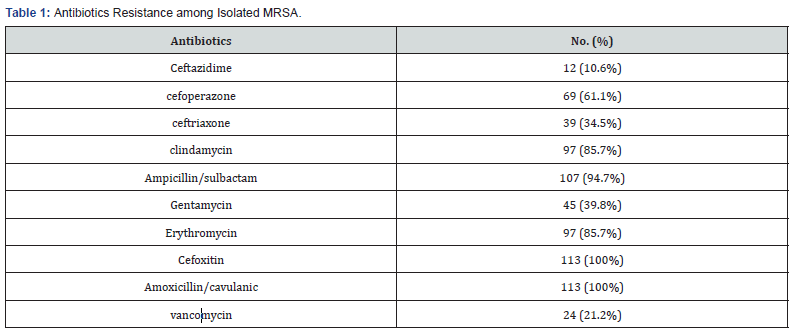
Results
The study included 365 isolated S.aureus strains. Among
isolated S.aureus strains, 113 (30.9%) was found to be MRSA,
Isolated MRSA species were all resistant tocefoxitin and amoxicillin/
clavulanic with high resistance to ampicillin/sulbactam (94.7%),
clindamycin and erythromycin (85.7%),vancomycin (21.2%).
Table 1.
Vancomycin resistance among MRSA according to MIC was
classified into susceptible, intermediate susceptible and resistant
according to CLSI. VRSA was 19 (16.8%), VISA was 10 (8.8%) with
MIC 4-8 μg/ml and susceptible strains were 84 (74.3%) with MIC
2μg/ml, data not shown.
Resistant vancomycin species was 15 strains with MIC 16-
32μg/ml, 3(2.7%) with MIC 64-128μg/ml, and one strain with
MIC 256μg/ml, while VRSA was found in 10 isolates (8.8%).
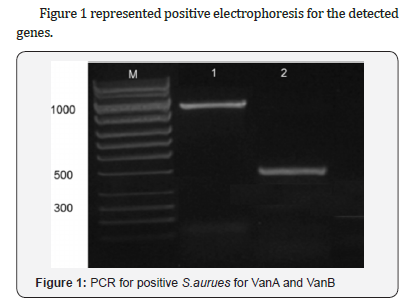
Van A gene was detected among 3 (30%) isolates with
intermediate susceptibility and in 13 (68.4%) resistant strains. vanB was more commonly associated with intermediate resistance
pattern in 6 (60%) isolates and in 17(89.5%) resistant strains and
none of the isolates had vanC, Table 2.

Discussion
The finding of the present study reported the presence of
MRSA in 30.9% among HAIs in children hospital during the period
of the study. The overall rates of MRSA in previous studies from
Egypt were up to 70% [11,12]. While lower rates were reported
in developed countries such as USA through implementing a
multi model intervention including active surveillance, contact
isolation, monitoring, and universal decolonization of patients in
intensive care units [13]. The difference between our results and
those from Egypt can be attributed to age of the included patients
and the sample size.
Our findings demonstrated high resistance of MRSA to betalactams
and macrolides antibiotics with rates from 85% up to
100%. These high rates of resistance are online by others reported
from other studies [14, 15]. The high rate of resistance could be
explained by the response of the MRSA strains to the selection
pressure created by their constant exposure to antibiotics used in
hospital settings [15].
In MRSA, 16.8% isolates were VRSA by determination of MIC
with different MIC ranging from 16 to 512 Mg/ml.
In Middle East countries various studies have reported the
presence of VRSA like Jordan [16], Saudi Arabia and Egypt [17].
In our study; about 20% of the isolates harbored at least one
of the van genes. There is a possibility that these infections were
caused by dissemination of a few clones of VRSA circulating in our
hospital but, we can neither confirm nor exclude this possibility
[18].
vanA gene was detected among 68.4% resistant strains and
vanB was detected among 89.5% of VRSA strains. Similarly,
vanA and vanB resistant genes were detected in 34% and 37%
of clinical isolates, respectively [18]. The absence of van genes
among VRSA strains are mainly due to the presence of other genes
and mechanisms that attribute to the emergence of these strains
in different proportions in VRSA
In this study, though we have found vanA and vanB genotypes
among VISA isolates with high frequency 30% and 60%
respectively. The presence of van genes A and B is considered
among other mechanisms of VISA like thickened cell wall [2].
Patients infected with these strains usually have resistant pattern
to vancomycin therapy when exposed to it. Moreover, the presence
of carrier for these strains can be a source for emergence of VRSA
isolates [19].
The findings highlight the emergence of vancomycin resistance
among methicillin resistant S.aureus isolated from children with
health care associated infections. Most resistant species revealed
the presence of vanA and vanB as a responsible mechanism for
this resistance.
Conclusion
The results of the current study illustrate the emergence
of vancomycin resistance among methicillin-resistant S. aureus
isolated from children with healthcare-associated infections.
The majority revealed the occurrence of vanA and vanB as an
accountable mechanism for this resistance.
To Know More About Trends in Technical and
ScientificResearch Please click on:
https://juniperpublishers.com/ttsr/index.php
To Know More About Open Access
Journals Please click on:




Comments
Post a Comment